SharkAI Guided Undergraduate Teaching Shark Segmentation (GUTSS) Application
Imagine diving into the fascinating world of sharks, where cutting-edge AI technology meets marine wonders. That's exactly what unfolded before my eyes as I stood before my soon-to-be McNair mentor.
Given my interest in learning more about marine life and my passion for exploring AI concepts, Dr. Waisome’s offer immediately piqued my interest. As a third-year Computer Science student desperately scrambling to find my next research opportunity, I gleefully took up the offer of having a new research mentor for the next year. With her recommendation in hand, I applied to Ronald E. McNair program, and, to my surprise, I made it in.
Given my fundamental ASV obstacle detection prototype that I created at Dartmouth Reality and Robotics Lab, I had background experience in AI. All I had to learn was shark dentistry, right?
I was dead wrong. As the spring semester came to an end, my mentor introduced me to Dr. Grant, the research director of the UF Data Studio. As he was also an associate professor on AI, ML, and data, he helped me establish my understanding of AI segmentation. Before the summer semester started, we already looked at Facebook’s SAM model, delving deep into Mixtec lore with figure segmentation. Particularly, we focused on the classification of figures of different codices once we isolated them through the Segment Anything Model (SAM).
With the SAM online demo, we isolated individually all the humans and creatures out of the pictures from ancient codices. Gradually, we learned how to make the process more streamlined and efficient, boxing several at once and then labeling the figures. Eventually, I gained an idea of what to do for the SharkAI project, based on the similarities in segmentation.
First, we needed a picture to test out how the SAM modeling will work:
 Figure 1 - This is our picture that we are going to run the SAM model on.
Figure 1 - This is our picture that we are going to run the SAM model on.
Next, we ran SAM through HiPerGator, UF’s supercomputer, over the picture, attempting to understand how each image token is cut out:
 Figure 2 - This is the result of running the picture through SAM Model Demo.
Figure 2 - This is the result of running the picture through SAM Model Demo.
With these first steps, we began extracting our ingredients for the AI tool, which will eventually be built to help the public learn about shark dissection. With what SAM describes, the main confounding parameters I learned were the IOU and stability thresholds [1]. With HuggingFace’s help, IOU impacts the prediction of the size of the area that the bounding box will highlight while stability influences the strength of the prediction [2].
In SAM, I started examining how each image layer works for each object segmentation, creating slices of each sector with different inputs to the model’s parameter until it has just the right number of masks or colors. In particular, I went through a tutorial on how to install and run SAM through HiPerGator.
Once I ran the system, I started to get my database, combing the Internet for pictures of teeth. Upon gathering the teeth photos, I divided them into three folders: simple, labeled, or unlabeled.
I followed how this table works:
| Category | Descriptions |
|---|---|
| Simple | Pictures with as little hues as possible and no texts and labels |
| Labeled | Pictures with text and labels and as much hues as possible |
| Unlabeled | Pictures with no text and labels and as much hues as possible |
After selecting my data, I ran it through SAM and changed the two parameters accordingly until it detected only the teeth or even parts of the teeth.
Here’s an example of the finale upon raising and lowering the parameters:
 Figure 3 - This is the result of running the picture through SAM Model on HiPerGator.
Figure 3 - This is the result of running the picture through SAM Model on HiPerGator.
However, we need to add the seasoning: the labeling to identify what the contents of each area of the picture are. With Grounded Dino, I eventually was able to identify the image components, whether it be teeth or spine. It will become the key part to let the students know what feature they will be looking at while taking the shark apart.
I started by installing and running Grounded Dino in HiPerGator. I used trial images that they give in order to make sure it runs properly. Then, I began using the old dataset to make sure that the text inputs work for its outputs.
Inside Grounding Dino’s parameters, I understood that the main components were the box and text inputs [3]. They were the ones that we control in order to understand what the user wants the algorithm to do as well as analyzing what identifier that we wanted to have.
For the shark teeth, I demonstrated what it would look like for identifying all the dental pieces in the picture:
 Figure 4 - This is the result of running the picture through Grounding DINO on HiPerGator.
Figure 4 - This is the result of running the picture through Grounding DINO on HiPerGator.
Being a programmer with minimal knowledge of shark anatomy, this process has also taught me about the in's and out's of sharks. Through SharkAI’s teacher conference, which was held at the Florida Museum for teachers from all across the state of Florida, we learned more about sharks and how they are usually taught across K-12 schools.
Eventually, our experience included a further exploration on how students learned all about anatomy from dogfish shark dissections. Therefore, I created a new dataset: one with dogfish shark anatomical figures like skeletons, organs, and other parts.
Here’s a little taste test of what to come:
 Figure 5 - Tagged unlabeled shark image from the web.
Figure 5 - Tagged unlabeled shark image from the web.
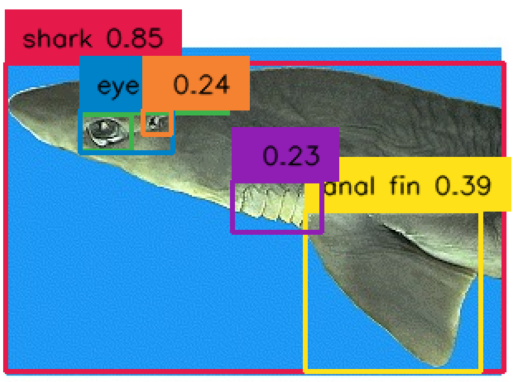 Figure 6 - Tagged labeled shark image.
Figure 6 - Tagged labeled shark image.
Hence, our stew -- Guided Undergraduate Training for Shark Segmentation (GUTSS)-- grew to a boil, which will gradually be doled out for marine biology learners to consume and learn in the upcoming year.
References
[1] A. Kirillov et al., “Segment Anything.” arXiv, Apr. 05, 2023. Accessed: Aug. 07, 2023. [Online]. Available: http://arxiv.org/abs/2304.02643
[2] “SAM,” huggingface.co. https://huggingface.co/docs/transformers/main/model_doc/sam (accessed Aug. 09, 2023).
[3] “🦕 Grounding DINO,” GitHub, May 08, 2023. https://github.com/IDEA-Research/GroundingDINO



